|
|
Transcription of DNA into RNA, enzymatic reactions, RNA, RNA degradation
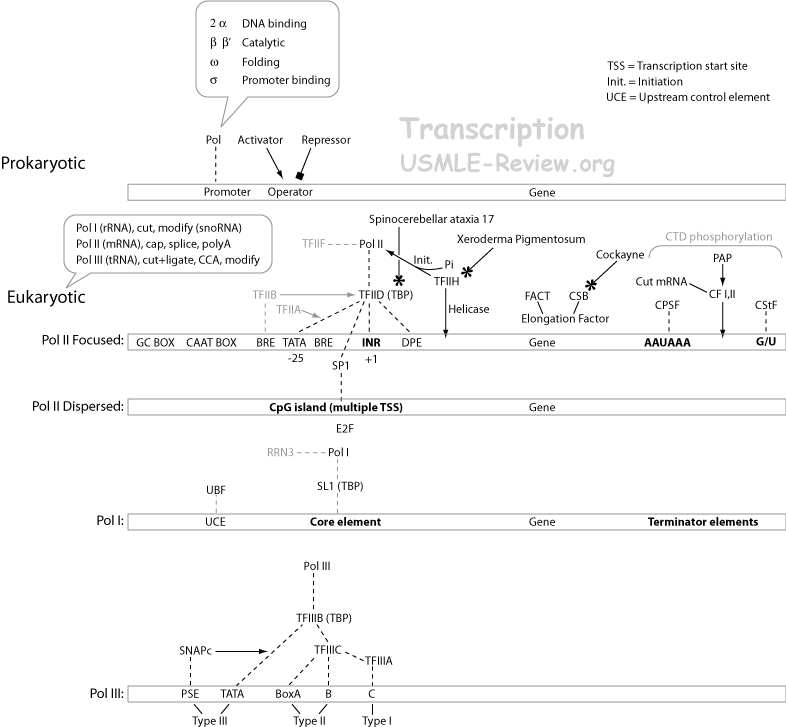
- Transcription
- Initiation: promoter recognition, closed complex, open complex.
- Promoter:
- Prokaryotic: ←upstream, -35 region, Pribnow box, transcription start site (TSS, +1), downstream→
- Eukaryotic: ←upstream, several upstream elements, TATA box, initiator element containing TSS (+1), downstream→
- The high A-T composition in promoters facilitate unwinding of DNA.
- Template strand = antisense strand = (-) strand = noncoding strand = the DNA strand that serves as the template for transcription.
- Nontemplate strand = sense strand = (+) strand = coding strand = the DNA strand having the same sequence as the transcribed RNA.
- Binding to promoter:
- Prokaryotic:
- holoenzyme = core enzyme (polymerase activity) + σ-subunit (promoter and strand specificity).
- binding first forms the closed complex, and then DNA opens up, forms the open complex.
- Eukaryotic:
- A whole bunch of transcription factors (TFs) involved in promoter recognition, binding, and openning up DNA.
- TBP = Tata binding protein. TAF = TBP associated factor.
- Phosphorylation of Pol II C-terminal domain (CTD) opens DNA up, forms the open complex.
- Polymerase must transcribe using the correct template strand. The σ-factor (prokaryotes) and TFs (eukaryotes) tell the RNA polymerase to bind the coding strand, while using the template strand as the template.
- Elongation:
- Polymerases:
- Prokaryotes have just one.
- Eukaryotes have three:
- RNA Pol I: makes rRNA (except the small 5S rRNA that resembles a tRNA in size).
- RNA Pol II: makes mRNA.
- RNA Pol III: makes tRNA (and 5S rRNA).
- Incorporation of NTPs.
- Prokaryotes lose σ-subunit. Eukaryotes lose TFs.
- Topoisomerases relaxing supercoils ahead and behind the polymerase.
- Transcription-coupled repair: RNA Pol II encounters DNA damage, backs up, TFIIH comes along, recruits repair enzymes. Defective TFIIH → faulty transcription-coupled repair → Xeroderma pigmentosum and Cockayne syndrome (skin sensitive to sunlight radiation in both diseases).
- Termination
- Prokaryotic:
- Intrinsic termination: GC hairpin (stalls polymerase) followed by poly U (slips off).
- Rho-dependent termination: ρ protein catches up to polymerase when it stalls at the hairpin, and bumps it off.
- Eukaryotic:
- Termination consensus sequence reached (AAUAAA).
- Polymerase released somewhere further downstream to the consensus sequence.
- RNA
- RNA = ribonucleic acid, has 2'-OH.
- rRNA = ribosomal RNA
- Most abundant (r for rampant).
- Catalyzes peptide bond formation in the ribosome.
- mRNA = messenger RNA
- Longest (m for massive).
- Contains sequence of codons for translation.
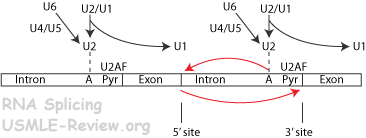
- RNA splicing
- pre-mRNA need to be processed.
- Introns = interfering sequences, cut out.
- Exons = spliced together.
- RNA splicing proceeds via a lariat intermediate, by the action of the spliceosome (snRNPs), introns released in lariat form.
- Some RNA can self splice.
- tRNA = transfer RNA
- Smallest (t for tiny).
- Contains anticodon.
- Shuttles the correct amino acid to the correct codon during translation.
- snRNPs (snurps) = RNA + protein, involved in RNA splicing.
- RNA degradation
- RNases degrade RNA.
- Post-transcriptional modifications protect RNA from degradation (5' cap and polyA tail)
- 2'-O-methylation prevents that position from attacking the RNA backbone.
Regulation: cis-regulatory elements, transcription factors, enhancers, promoters, silencers, repressants,
splicing
- Prokaryotic:
- Operons:
- Inducible operon (lac operon): normally repressed (operator bound by LacI's repressor protein), but is induced (repressor inactivated) in the presence of an effector molecule (allolactose). Lac operon structure: LacI-Promoter-Operator-Lac genes.
- Repressible operon (trp operon): normally functioning (trpR makes inactive repressor), but is repressed (repressor activated and binds to operator) in the presence of an effector molecule (trp). Trp operon structure: trpR-----Promoter-Operator-Trp genes.
- Repressors inhibit transcription when bound to regulatory sequence near promoter (eg. LacI on operator of the lac operon).
- Activators activate transcription when bound to regulatory sequence near promoter (eg. CAP-cAMP on CAP site in promoter of the lac operon).
- An operon can regulate several downstream genes by transcribing a polygenic mRNA (eg. LacZ, lacY, lacA).
- Attenuation:
- Attenuation sequence within Trp gene: TrpL-Pause site-Antiterminator-Terminator.
- Pause site: contains a lot of trp codons. If there's not a lot of trp amino acids around, it will cause the ribosome to pause.
- If paused (cell starved of trp), antiterminator forms, and the trp mRNA is translated.
- If not paused (cell has lots of trp), terminator forms, and translation terminates.
- Eukaryotic:
- Operons are rare.
- Chromatin configuration:
- Histone binding to DNA represses genes.
- Activators
- Bind to promoter elements and enhancers.
- Binding to promoter elements occur near the promoter.
- Enhancer-binding elements bind to enhancers (sites remote from the promoter, but influence it by looping back onto it).
- Helps recruit polymerase.
- Activate genes by blocking histone binding.
- Repressors: Eukaryotes are less concerned with repressors because chromatin structure (histones-binding) is repressive. Though, some exist:
- Bind to DNA adjacent to activator, and inactivates it.
- Binds to the same site as activator, and blocks it.
- Recruit histone deacetylases.
- Silencers: an enhancer that is bound by a strong repressor, which can overcome (silence) activators binding to promoter elements.
- Chromatin remodelling:
- Histone acetylation (activate gene) and deacetylation (reverses acetylation).
- Nucleosome remodelling complex: slides nucleosomes so different parts of DNA are exposed/bound.
- Gene silencing:
- Histone deacetylation.
- CpG methylation.
- Imprinting:
- Prader-Willi syndrome: inactivation or deletion of paternal genes on chromosome 15. Symptoms: small + weak at birth, mental retardation, after 12 months develops uncontrollable hunger/eating, obesity, diabetes.
- Angelman syndrome: inactivation or deletion of maternal genes on chromosome 15. Symptoms: mental retardation, motor retardation, small head size, hyper activity, inappropriate laughter.
- Telomere position effect: active genes moved to the telomere region gets silenced.
- RNAi: small dsRNA that matches sequence of a gene will recruit protein complex (RISC = RNA-induced silencing complex) that silences the gene.
- RNA processing:
- 5'-cap
- 3'-polyA tail: the longer the tail, the longer the mRNA's life, the more its translated.
- Polyadenylation enzyme adds 3'-polyAs.
- Deadenylation enzyme removes 3'-polyAs.
- RNA splicing.
- RNA with splice junctions and branch-point adenine recruits proteins (snRNPs) that make up the spliceosome.
- Introns cut out and released as a lariat. Exons joined back together.
- Alternative splicing: different ways to splice an RNA makes different mRNA products.
- RNA editing inserts, deletes, or change nucleotides in an RNA.
- RNA transport: mRNA that gets out of the nucleus gets transcribed. Those that stay in the nucleus gets degraded.
- mRNA degradation: controls lifetime of mRNAs.
- Deadenylation enzyme 3'-polyA tail.
- Decapping enzyme removes 5'-cap.
- Exonucleases and endonucleases.
- Cis-regulatory elements (aka cis-acting elements): regulatory elements on the same DNA molecule as the gene they regulate. Do not produced diffusible elements that can act on other DNA.
- Operator in operons.
- Promoter
- strong core promoter consensus sequence means more transcription.
- promoter proximal elements:
- House-keeping genes have promoter proximal elements that are bound by ubiquitous activators.
- Cell-specific genes have promoter proximal elements that are bound by cell-specific activators.
- Enhancer: usually upstream of the gene. Activators and protein complexes bind to it, DNA loops back to bring the enhancer with all the attached proteins in close proximity to the transcription site.
- Trans-regulatory elements: genes that make activators/repressors proteins (diffusible elements) which act on a different strand of DNA. Eg. LacI.
|
|