|
|
the genetic code
- The genetic code: converts mRNA codons into amino acids.
- Codons are triplets: coded by 3 nucleotides.
- Start codon: AUG (Augment means to increase), codes for methionine.
- Stop codons: UGA, UAA, UAG (U Go Away, U Are Away, U Are Gone)
- Commaless: read without gaps.
- Nonoverlapping.
- Universal: almost all organisms use the same code.
- Degenerate: More than one codon per amino acid.
- Wobble hypothesis: molecular mechanism for codon degeneracy is that the third letter of the codon can wobble in its bonding with the anticodon.
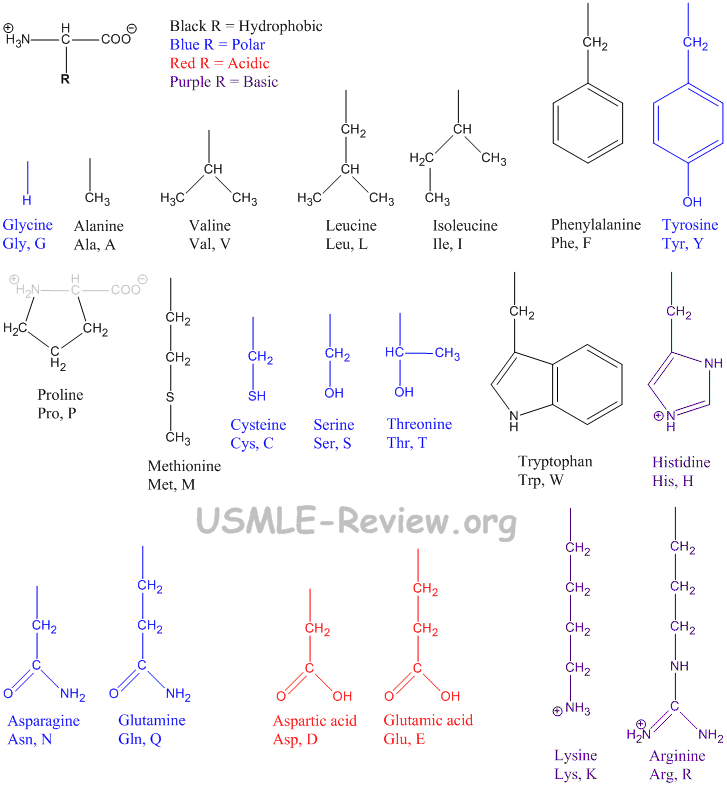
- Amino Acids
structure and function of tRNA
- tRNA structure: cloverleaf secondary structure, L-shaped tertiary structure.
- Tip contains the anticodon.
- 3'- ends in CCA: the amino acid is attached to the 3'-OH of the A.
- tRNA function: bring the correct amino acid to the correct codon.
- Charging a tRNA: aminoacyl-tRNA synthetase attaches the correct amino acid to the correct tRNA. Specificity determined by tRNA structure interaction with aminoacyl-tRNA synthetase.
- tRNA brings the correct amino acid to the correct codon. Specificity determined by codon-anticodon interaction.
structure and function of ribosomes
- Ribosome structure:
- Prokaryotic: 70S = 30S + 50S
- Small subunit: 30S = 16S rRNA + proteins.
- Large subunit: 50S = 23S rRNA + proteins + 5S rRNA.
- Eukaryotic: 80S = 40S + 60S
- Small subunit: 40S = 18S rRNA + proteins.
- Large subunit: 60S = 28S rRNA + proteins + 5S rRNA + 5.8S rRNA.
- Ricin/abrin/α-sarcin inactivates 28S rRNA.
- Ribosome function: protein synthesis
protein synthesis
- Prokaryotic:
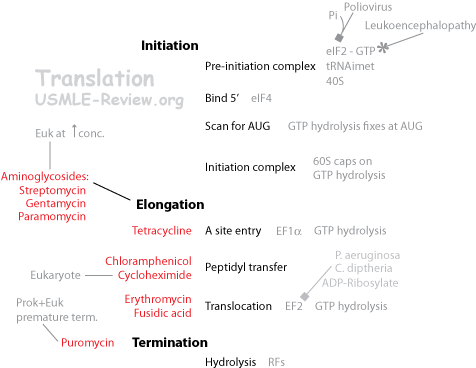
- Initiation:
- Shine-Dalgarno sequence on mRNA bind small subunit. tRNA f-met bind start codon on mRNA at the P site.
- Initiation factors (IFs) help bring all the above together. IF-1 and IF-3 helps mRNA binding. IF-2 + GTP helps tRNA binding.
- IFs dissociate, large subunit comes along and caps over the small subunit, sandwiching mRNA and tRNA in between.
- Aminoglycosides affect initiation and causes mRNA misreading.
- Paromomycin: increases error rate.
- Streptomycin: inhibits initiation.
- Elongation:
- Decoding: aminoacyl-tRNA's anticodon matches mRNA codon at the A site. EF-Tu + GTP help bring aminoacyl-tRNA to the A site. EF-Ts recharges EF-Tu with GTP.
- Tetracycline inhibits aminoacyl-tRNA binding.
- Transpeptidation: Peptide bond forms from the N-term of A site amino acid attacks the C-term of the P site chain. Results in chain migrated to A site. The reaction is catalyzed by the rRNA of the large subunit.
- Puromycin is an analogue of tyr-tRNA with a dead C-term (amide rather than ester), causes premature termination.
- Chloramphenicol inhibits peptidyl transferase in prokaryotic large subunit.
- Translocation: the tRNAs along with the mRNA moves down the ribosome. The previous P site tRNA enters E site (exits), and the previous A site tRNA (holding the chain) moves to the P site. The A site is now empty and ready to accept the next tRNA. EF-G + GTP needed.
- Erythromycin inhibits prokaryotic translocation in large subunit.
- Fusidic acid affects EF-G (prevents it from dissociating from large subunit).
- Macrolides bind 50S, inhibit translocation.
- Clindamycin binds 50S, inhibit translocation.
- Termination:
- Release factor 1 (RF1) or RF2 binds stop codon.
- Hydrolysis of ester bond between C-term of protein chain and 3'-tRNA.
- RF3-GTP knocks off RF1 or RF2, then dissociates itself.
- Ribosomal recycling factor (RRF) and EF-G + GTP knocks off the tRNA, the mRNA, and then dissociate themselves.
- Eukaryotic: similar to prokaryotic, with the most difference in initiation.
- Initiation:
- initator tRNA-met binds to small subunit by initiation factors at the P site.
- 5'-cap and 5'-UTR of mRNA binds to small subunit by initiation factors.
- tRNA anticodon slides along mRNA until it finds AUG start codon.
- IFs dissociate, large subunit comes along and caps over the small subunit, sandwiching mRNA and tRNA in between.
- Eukaryotes have a whole lot more IFs than prokaryotes.
- Elongation:
- Decoding: aminoacyl-tRNA's anticodon matches mRNA codon at the A site. eEF1A (Eukaryotic equivalent of EF-Tu) and eEF1B (Eukaryotic equivalent of EF-Ts) needed.
- Transpeptidation: formation of peptide bond, where A site amino acid N-term attacks P site chain's C-term.
- Puromycin mimics tyr-tRNA but has a dead C-term, and causes premature termination because transpeptidation can't occur.
- Cycloheximide inhibits peptidyl transferase on eukaryotic large subunit.
- Translocation: tRNA, mRNA slides down ribosome. P (tRNA) → E, A (tRNA + chain) → P, A site now empty. eEF2 (Eukaryotic equivalent of EF-G) needed.
- Diphtheria toxin inactivates eEF2.
- Termination:
- eRF1 (Eukaryotic release factor) binds stop codon.
- Hydrolysis of ester bond between C-term of protein chain and 3'-tRNA.
- eRF3 (Eukaryotic equivalent of RF3) knocks off eRF1, then dissociates.
- Eukaryotic equivalent of RRF and eEF2 involved in knocking off mRNA and tRNA (not well understood).
regulation of translation
- Regulation of gene expression in prokaryotes is predominantly transcriptional control. Although, differences in Shine-Dalgarno sequences play a minor role in translational control.
- Eukaryotes have regulation of translation because their mRNA have a much longer life time.
- Eukaryotic initiation factors.
- Reticulocytes translate hemoglobin when there's heme present (normal eIF2), but stops translating hemoglobin when heme is scarce (phosphorylated eIF2).
- Heme-regulated inhibitor (HRI) is the kinase.
- eIF2 phosphatase reverses the action of HRI.
- Interferons inhibits translation of viral proteins.
- Interferons induce an eIF2 kinase (PKR/DAI), which is activated upon binding to viral dsRNA. Many viruses express PKR inhibitors.
- Interferons also induce mRNA degradation (RNase L activated by presence of dsRNA).
- PKR-like endoplasmic reticulum kinase (PERK) phosphorylates eIF2 when there's too much unfolded proteins in the ER. This prevents the cell from being overwhelmed and damaged.
- Wolcott-Rallison syndrome: defective PERK leads to pancreatic β cells, and causes type I diabetes and multiple systemic disorders.
- Cap-binding protein (eIF4E)
- More active when phosphorylated.
- Binding of inhibitors inactivates cap-binding proteins.
- mRNA masking: binding of mRNA to proteins prevent them from being translated.
- mRNA in oocyte is masked by formation of ribonucleoporotein particles (RNPs).
- Regulate gene expression in early development by unmasking the stored RNPs.
- Cytoplasmic polyadenylation
- Normally, polyA is added in nucleus, but cytoplasmic polyA addition regulates translation on another level.
- Antisense oligonucleotides:
- Antisense hybridizes with mRNA, forms dsRNA that is target for RNase H.
- RNase H cleaves mRNA but leaves antisense (chemically modified, resistant to RNase)
- Antisense can go hybridize again.
post-translational modifications, including phosphorylation, addition of CHO units
- Cleavage:
- Remove initiator met or f-met.
- Remove signal peptide (initially used to target the RER).
- Zymogen (proenzyme) → active enzyme.
- -sinogen → -sin. Eg. Pepsinogen → pepsin.
- Procollagen → collagen
- Ehlers-Danlos syndrome VII: mutation in procollagen that prevents it from being cleaved.
- Polyproteins → protein components.
- Modification:
- Hydroxylation of procollagen chain so that the mature collagen triple helix can hydrogen bond.
- Glycosylation to make glycoproteins by adding sugar residues.
- Phosphorylation of mannose targets protein to lysosomes.
- Complex carbohydrates targets protein to cell membrane.
- Dolichol transfers initial carbohydrate chain onto emerging peptide. Then all but one glucose residue (recognized by chaperones) is removed.
- Chaperones like calreticulin (CRT), calnexin (CNX), and BiP prevent aggregation of unfolded glycoprotein. Misfolded proteins are degraded.
- Phosphorylation to activate or inactivate enzymes.
- Acetylation. Eg. Histone acetylation activates genes.
- Methylation. Eg. Histone methylation restricts histone deacetylation.
- Splicing:
- Intein cut out.
- Exteins spliced together.
- Inteins are homing endonucleases, cut inteinless extein genes, then insert intein gene (like molecular parasite, mechanism of insertion is by recombination with homologous intein-containing extein gene).
protein degradation
- Targets for degradation:
- Random degradation responsible for turnover of proteins.
- Abnormal proteins are degraded.
- Proteins that are involved in cell-cycle control (eg. cyclins).
- Protein degradation goes up when cell is starved to provide energy.
- Mechanisms:
- Ubiquitin tags protein for degradation.
- Ubiquitin (analogous to C-term) forms amide bond with N-term of protein.
- E1, E2, E3 helps ubiquitin to bond with protein.
- Protein degraded inside the proteosome (barrel) with ATP hydrolysis.
- Ubiquitin is not degraded by the proteosome.
- Lysosomes: fusion with either autophagic vacuoles or with endocytosed vacuoles.
- Diabetes mellitus stimulates lysosomal degradation of proteins.
- Muscle wastage.
- Rheumatoid arthritis.
|
|